BNPgraph package: demo_experiments
This Matlab script performs posterior inference on the yeast protein interaction network.
For downloading the package and information on installation, visit the BNPgraph webpage.
Reference: F. Caron and E.B. Fox. Sparse graphs using exchangeable random measures. arXiv:1401.1137, 2014.
Author: François Caron, University of Oxford
Tested on Matlab R2014a.
Contents
General settings
clear all % Add paths addpath('./GGP/'); addpath('./utils/'); set(0,'DefaultAxesFontSize',14) % Set the seed rng('default')
Load data
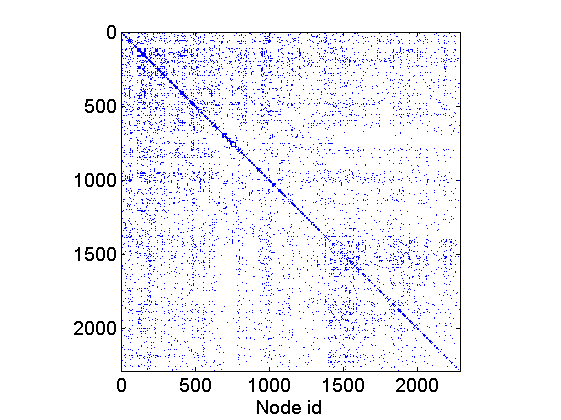
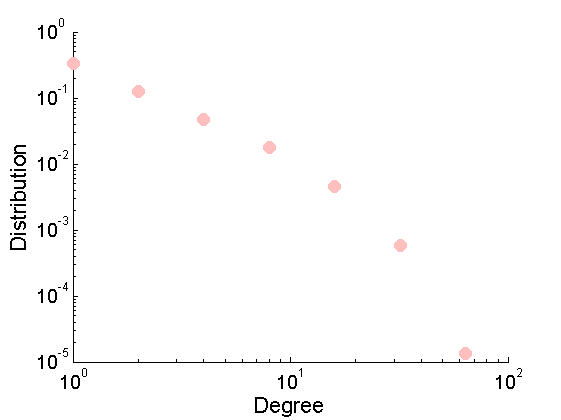
% Download the mat file from the pajek website namefile = 'yeast.mat'; urlwrite('http://www.cise.ufl.edu/research/sparse/mat/Pajek/yeast.mat',namefile); % Load the data load(namefile); G = Problem.A; % Remove self-edges and nodes with no connection G = G-diag(diag(G)); ind =(sum(G,1) + sum(G,2)')>0; G = sparse(logical(G(ind, ind))); % Plot adjacency matrix figure spy(G) xlabel('Node id'); % Plot empirical degree distribution figure h2 = plot_degree(G); set(h2, 'markersize', 10, 'marker', 'o',... 'markeredgecolor', 'none', 'markerfacecolor', [1, .75, .75]) box off
Posterior inference
% Parameters of the model hyper_alpha =[0,0]; hyper_tau = [0,0]; hyper_sigma = [0, 0]; objprior = graphmodel('GGP', hyper_alpha, hyper_sigma, hyper_tau, 'simple'); % Parameters of the MCMC algorithm niter = 40000; nburn = 0; nadapt = niter/4; thin = 20; nchains = 3; store_w = false; verbose = false; % Run MCMC tic objmcmc = graphmcmc(objprior, niter, nburn, thin, nadapt, nchains, store_w); % Create a MCMC object objmcmc = graphmcmcsamples(objmcmc, G, verbose); % Run MCMC time = toc;
-----------------------------------
MCMC chain 1/3
-----------------------------------
Start MCMC for GGP graphs
Nb of nodes: 2284 - Nb of edges: 6646
Number of iterations: 40000
Estimated computation time: 0 hour(s) 5 minute(s)
Estimated end of computation: 01-May-2015 13:52:10
-----------------------------------
-----------------------------------
End MCMC for GGP graphs
Computation time: 0 hour(s) 2 minute(s)
End of computation: 01-May-2015 13:49:10
-----------------------------------
-----------------------------------
MCMC chain 2/3
-----------------------------------
Start MCMC for GGP graphs
Nb of nodes: 2284 - Nb of edges: 6646
Number of iterations: 40000
Estimated computation time: 0 hour(s) 2 minute(s)
Estimated end of computation: 01-May-2015 13:50:52
-----------------------------------
-----------------------------------
End MCMC for GGP graphs
Computation time: 0 hour(s) 2 minute(s)
End of computation: 01-May-2015 13:50:51
-----------------------------------
-----------------------------------
MCMC chain 3/3
-----------------------------------
Start MCMC for GGP graphs
Nb of nodes: 2284 - Nb of edges: 6646
Number of iterations: 40000
Estimated computation time: 0 hour(s) 2 minute(s)
Estimated end of computation: 01-May-2015 13:52:50
-----------------------------------
-----------------------------------
End MCMC for GGP graphs
Computation time: 0 hour(s) 2 minute(s)
End of computation: 01-May-2015 13:52:30
-----------------------------------
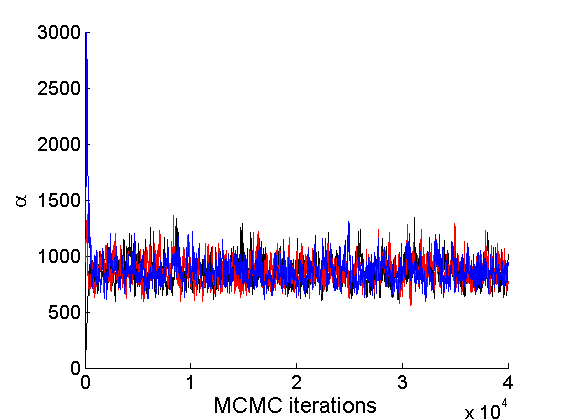
Trace plots and posterior histograms
nsamples = length(objmcmc.samples(1).w_rem); [samples_all, estimates] = graphest(objmcmc, nsamples/2) ; % concatenate chains and returns estimates col = {'k', 'r', 'b'}; % Trace plots variables = {'alpha', 'sigma', 'tau', 'w_rem'}; names = {'\alpha', '\sigma', '\tau', 'w_*'}; for j=1:length(variables) figure for k=1:nchains plot(thin:thin:niter, objmcmc.samples(k).(variables{j}), 'color', col{k}); hold on end legend boxoff xlabel('MCMC iterations', 'fontsize', 16); ylabel(names{j}, 'fontsize', 16); box off end
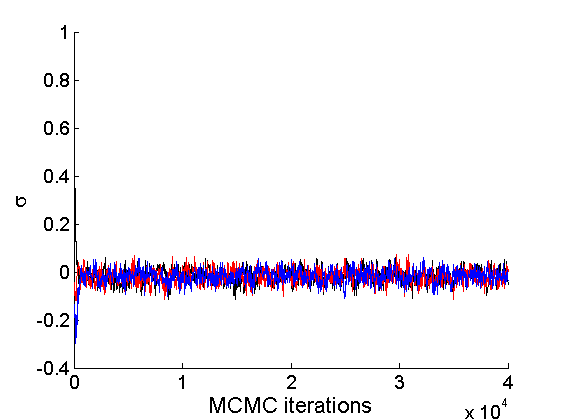
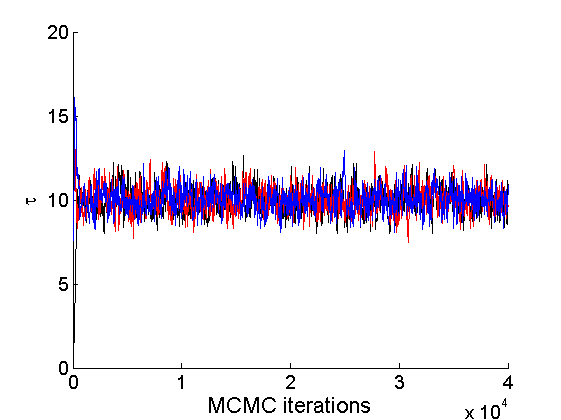
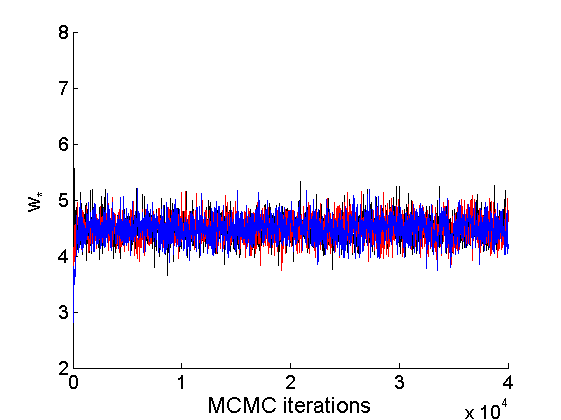
% Posterior Histograms variables = {'alpha', 'sigma', 'tau', 'w_rem'}; names = {'\alpha', '\sigma', '\tau', 'w_*'}; for j=1:length(variables) figure hist(samples_all.(variables{j}), 30); legend boxoff ylabel('Number of samples', 'fontsize', 16); xlabel(names{j}, 'fontsize', 16); box off end
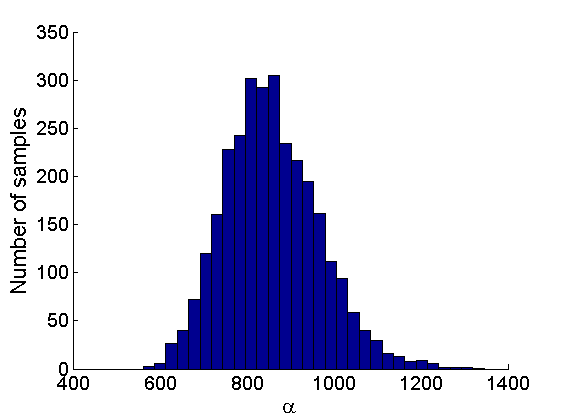
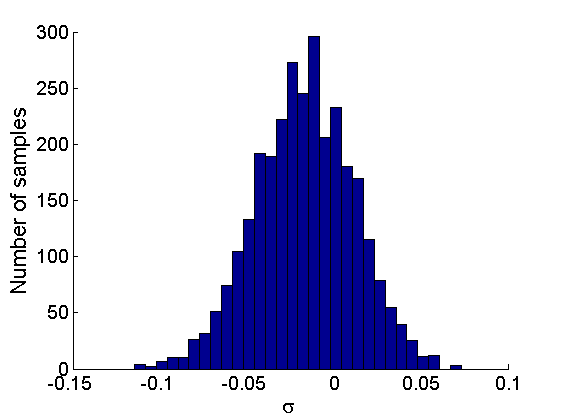
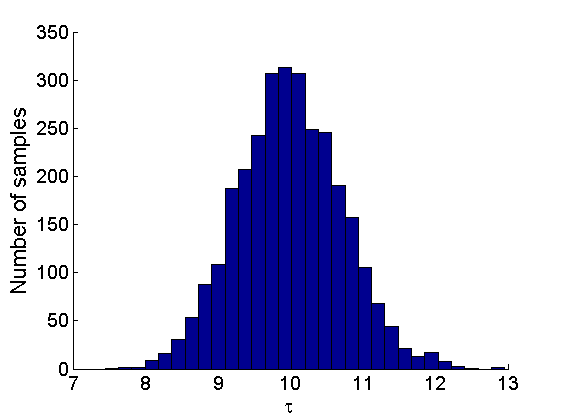
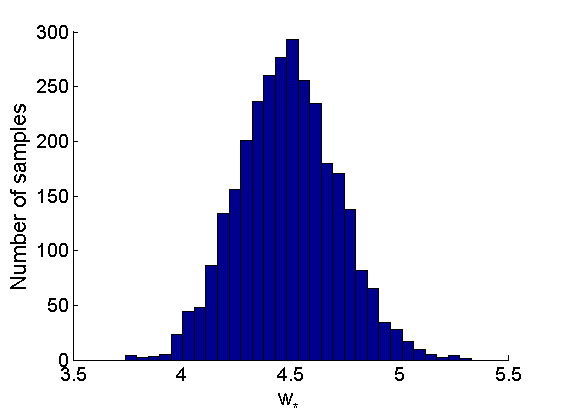
Assessing the sparsity of the network
% Probability that sigma>=0 proba_sparse = mean(samples_all.sigma>=0); fprintf('Probability of sparse graph = %.3f \n', proba_sparse); fprintf('99 %% posterior interval for sigma = [%.3f,%.3f] \n',... quantile(samples_all.sigma, [.005, .995]));
Probability of sparse graph = 0.280 99 % posterior interval for sigma = [-0.093,0.054]
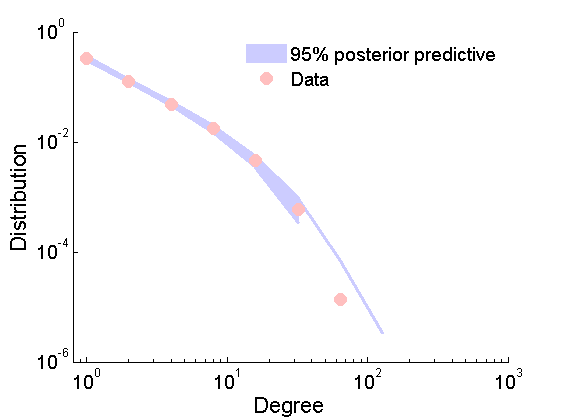
Posterior predictive degree distribution
% Posterior predictive degree distribution nsamples_all = length(samples_all.alpha); ndraws = 2000; ind =floor(linspace(1,nsamples_all,ndraws)); freq = zeros(ndraws, 13); htemp=figure('Visible', 'off'); for ii=1:ndraws % Samples from the predictive if rem(ii, 200)==0 fprintf('Sample %d/%d from the posterior predictive\n', ii, ndraws); end Gsamp = graphrnd(graphmodel('GGP', samples_all.alpha(ind(ii)), samples_all.sigma(ind(ii)), samples_all.tau(ind(ii))), 1e-6); [h2, centerbins, freq(ii, :)] = plot_degree(Gsamp); end close(htemp); plot_variance = @(x,lower,upper,color) fill([x,x(end:-1:1)],[upper,lower(end:-1:1)],color, 'EdgeColor', color);%set(,'EdgeColor',color); quantile_freq = quantile(freq, [.025, .975]); figure plot(centerbins, quantile_freq, 'color', [.8, .8, 1], 'linewidth', 2.5); hold on ind2 = quantile_freq(1,:)>0; ha = plot_variance(centerbins(ind2), quantile_freq(1,ind2),quantile_freq(2,ind2), [.8, .8, 1] ); set(gca,'XScale','log') set(gca,'YScale','log') hold on hb = plot_degree(G); set(hb, 'markersize', 10, 'marker', 'o',... 'markeredgecolor', 'none', 'markerfacecolor', [1, .75, .75]) legend([ha, hb],{'95% posterior predictive', 'Data'}) legend boxoff xlim([.8, 1e3]) box off
Sample 200/2000 from the posterior predictive Sample 400/2000 from the posterior predictive Sample 600/2000 from the posterior predictive Sample 800/2000 from the posterior predictive Sample 1000/2000 from the posterior predictive Sample 1200/2000 from the posterior predictive Sample 1400/2000 from the posterior predictive Sample 1600/2000 from the posterior predictive Sample 1800/2000 from the posterior predictive Sample 2000/2000 from the posterior predictive