Department of Statistics
24–29 St Giles'
Oxford OX1 3LB

My research explores the potential of computational statistics and statistical machine learning to assist in the medical and health sciences. In this respect I oversee a small research group working on probabilistic models and Bayesian decision analysis in complex biomedical data environments. This includes theoretical foundations, novel methodology, and “hands-on” study driven data science.
I hold a joint Statutory Professorship (Oxford speak for Chair) in Biostatistics at the departments of Statistics and the Nuffield Department of Medicine. Within the Nuffield medical school I am an Affiliate Member of the Li Ka Shing Centre for Health Information and Discovery. My research is partly funded through a Programme Leaders award in Statistical Genomics from the UK's Medical Research Council. I am Scientific Director for the Health Programme at the Alan Turing Institute, London.

The University of Oxford Big Data Institute has recently engaged in a long-term research collaboration with Novartis. The aim of the collaboration is to develop statistical machine learning methods to better understand complex diseases using Novartis' clinical trial data. The partnership will initially focus on flagship programs in multiple sclerosis (MS), rheumatology and dermatology. MS is a chronic and ultimately debilitating disease that affects approximately 2.5 million individuals worldwide. A disease of the central nervous system, MS is characterised by the inflammation and eventual destruction of the axons. Novartis has amassed a vast database from clinical trials targeting MS, including brain MRI images across multiple modalities, clinical and genomic data. Using this data, the project aims to better characterise MS over the span of the disease and find biomarkers for early diagnosis, monitoring and prognosis of individual MS patients. The rheumatology and dermatology program will focus on the following autoimmune disorders: ankylosing spondylitis, rheumatoid arthritis, psoriatic arthritis and psoriasis. The goal of the study is to analyse the relevant studies and identify new factors driving disease progression and therapeutic response and develop cutting-edge tools to support clinical decision making.
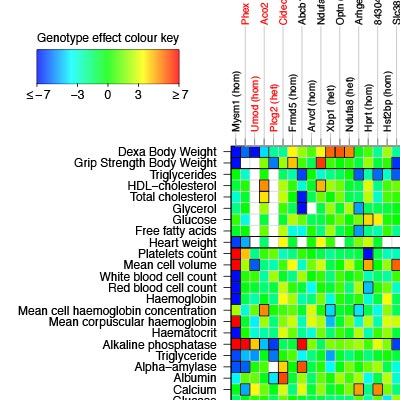
This project aims to develop and apply computational statistics and machine learning methods to enhance interpretation of data from the International Mouse Phenotyping Consortium and to facilitate their use in identification of models for human disease. The IMPC is a multi-centre collaboration aimed at measuring the phenotypic consequences of knocking out each gene in the mouse genome in turn. Several hundred measurements are taken on each animal, in procedures ranging from clinical blood chemistry, through calorimetry and body composition to behavioural phenotypes. Our current focus is on the use of sparse hierarchical factor models to effectively identify and interpret multivariate phenotype perturbations and impute unmeasured phenotypes.
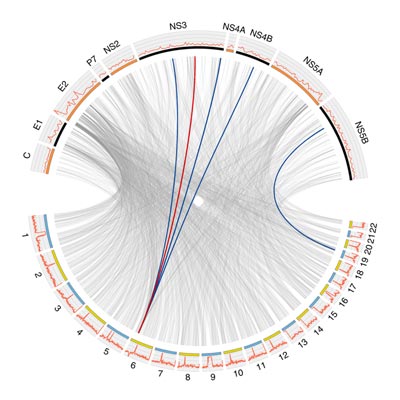
HCV infects around 200,000 people within the UK and 200 million people worldwide (2% of the world population). STOP-HCV is a flexible and dynamic UK wide consortium that will use patient stratification to optimise treatment of infected patients. The consortium builds on existing cutting edge clinical and scientific expertise, in partnership with industry. Our overarching aim is to define and develop a deeper understanding of patient strata and to develop prognostic models so that rational treatment strategies can be deployed. In a new era of novel Directly Acting Antiviral (DAA) therapies, treating only a subset of patients with DAA will cost the NHS an estimated £96 million/year. Therefore, refined patient stratification will be of enormous clinical and economic benefit. A focus of our program will be study of HCV genotype-3 infection, highly prevalent in the UK, with a characteristic clinical phenotype, and a higher relapse rate with DAA therapy. We will also focus on difficult-to-treat patient groups such as those with cirrhosis and those co-infected with HIV, where optimal management pathways will be of particular benefit in patients. Our consortium is underpinned by HCV Research UK, a network of 18 UK centres biobanking samples from 10,000 HCV infected patients, linked to a state-of-the-art clinical database. A unique aspect of the STOP-HCV consortium is the availability of complementary datasets on a common set of samples. This allows for integrative analysis, using all of the data for improved scientific understanding of the mechanisms underlying heterogeneity of disease susceptibility and progression, as well as host-viral interaction and implications for therapeutic response. The joint data will also enable an integrative approach to biomarker panel construction using genetic, genomic, and serum marker data, informed by the in vitro immunity experiments. Integrative analysis will take place in phases beginning with pair-wise analysis of datasets prior to integration of heterogeneous sources of data.

The aim of the combined effort of S-CORT consortium is to better diagnose colorectal cancer (CRC) in such a way as to increase the likelihood that the treatment with the highest chance of success, is prescribed to patients. It also aims to minimise the potential negative side effects associated with various therapies. Part of this work is to develop novel statistical methods, employing computational statistics and machine learning approaches, in biostatistics and statistical genomics to integrate the multi-omics data (DNA sequence, methylation, transcriptome and patient records) generated by the consortium in order to provide a greater biological understanding of CRC and how that underlies the prediction of outcome.

New technologies are providing opportunities to measure health and disease in many novel ways. This project focuses on two such technologies that measure genome-wide gene expression (RNA-seq transcriptomics) and concentrations of a broad range of small molecules involved in metabolism (metabolomics). One of the next big steps in precision medicine promises to be the integration of genetic data with such longitudinally varying molecular phenotypes to enhance prediction of disease and stratify treatments across patient groups to improve health outcomes. This project gathers RNA-Seq and metabolomic data longitudinally from ~700 members of the TwinsUK cohort, with these data accompanied by genotypes and extensive clinical and lifestyle information. We will explore how molecular traits track and vary over time, determine how such variation relates to underlying genetic variation, and explore the joint contribution of genetic and genomic data to disease risk, with a particular focus on type II diabetes. We will develop bespoke statistical and machine learning approaches to infer the longitudinal multivariate relationships amongst genotype, molecular traits, and environmental/lifestyle factors; and their application to identify robust, reproducible signatures associated with disease susceptibility and onset.
We are members of the Computational Statistics and Machine Learning group within the Department of Statistics.
Projects and group funded by:
| Title | Year | Research Area |
|---|---|---|
|
On the marginal likelihood and cross-validation E Fong, C Holmes Biometrika (to appear) |
2019 |
|
|
treeSeg: testing for dependence on tree structures M Behr, M A Ansari, A Munk, C Holmes bioRxiv, 622811 |
2019 |
|
|
Scalable Nonparametric Sampling from Multimodal Posteriors with the Posterior Bootstrap E Fong, S Lyddon, C Holmes ICML (2019) |
2019 |
|
|
Nonparametric learning from Bayesian models with randomized objective functions S Lyddon, S Walker, C Holmes NeurIPS (2018): Advances in Neural Information Processing Systems 31 |
2018 |
|
|
General Bayesian Updating and the Loss-Likelihood Bootstrap S Lyddon, C Holmes, S Walker |
2018 |
|
|
A Framework for Adaptive MCMC Targeting Multimodal Distributions E Pompe, C Holmes, K Łatuszyński arXiv preprint arXiv:1812.02609 |
2018 |
|
|
Transcriptome Deconvolution of Heterogeneous Tumor Samples with Immune Infiltration Z Wang, S Cao, JS Morris, J Ahn, R Liu, S Tyekucheva, F Gao, B Li, W Lu, ... iScience 9, 451-460 |
2018 |
|
|
Semi-unsupervised Learning of Human Activity using Deep Generative Models M Willetts, A Doherty, S Roberts, C Holmes arXiv preprint arXiv:1810.12176 |
2018 |
|
|
Probabilistic Boolean Tensor Decomposition T Rukat, C Holmes, C Yau International Conference on Machine Learning, 4410-4419 |
2018 |
|
|
Principles of bayesian inference using general divergence criteria J Jewson, J Smith, C Holmes Entropy 20 (6), 442 |
2018 |
|
|
M, S Hollowell, L Aslett, C Holmes, A Doherty Nature Scientific Reports, 8 |
2018 |
|
|
TensOrMachine: Probabilistic Boolean Tensor Decomposition T Rukat, CC Holmes, C Yau To appear, ICML (2018): arXiv preprint arXiv:1805.04582 |
2018 |
|
|
High-throughput mouse phenomics for characterizing mammalian gene function SDM Brown, CC Holmes, AM Mallon, TF Meehan, D Smedley, S Wells Nature Reviews Genetics, 1 |
2018 |
|
|
NOX1 loss-of-function genetic variants in patients with inflammatory bowel disease T Schwerd, RV Bryant, S Pandey, M Capitani, L Meran, JB Cazier, J Jung, ... Mucosal immunology 11 (2), 562 |
2018 |
|
|
Interferon lambda 4 impacts broadly on hepatitis C virus diversity MA Ansari, E Aranday-Cortes, CLC Ip, A da Silva Filipe, LS Hin, ... bioRxiv, 305151 |
2018 |
|
|
Multiscale blind source separation M Behr, C Holmes, A Munk The Annals of Statistics 46 (2), 711-744 |
2018 |
|
|
Principled Bayesian Minimum Divergence Inference J Jewson, JQ Smith, C Holmes Special Edition on the Foundations of Statistics, Entropy, 20(6), 442 |
2018 |
|
|
A general framework for predicting the transcriptomic consequences of non-coding variation M Abdalla, MI McCarthy, CC Holmes bioRxiv, 279323 |
2018 |
|
|
A Doherty, K Smith-Bryne, T Ferreira, MV Holmes, C Holmes, SL Pulit, ... bioRxiv, 261719 |
2018 |
|
|
QF Wills, E Mellado-Gomez, R Nolan, D Warner, E Sharma, J Broxholme, ... BMC genomics 18 (1), 53 |
2017 |
|
|
Generalized Bayesian Updating and the Loss-Likelihood Bootstrap S Lyddon, C Holmes, S Walker arXiv preprint arXiv:1709.07616 |
2017 |
|
|
Adaptive MCMC for multimodal distributions C Holmes, K Łatuszyński, E Pompe |
2017 |
|
|
AC Daly, J Cooper, DJ Gavaghan, C Holmes Journal of The Royal Society Interface 14 (134), 20170340 |
2017 |
|
|
Better together? Statistical learning in models made of modules PE Jacob, LM Murray, CC Holmes, CP Robert arXiv preprint arXiv:1708.08719 |
2017 |
|
|
J Watson, L Nieto-Barajas, C Holmes Statistics 51 (3), 558-571 |
2017 |
|
|
Principles of Experimental Design for Big Data Analysis CC Drovandi, C Holmes, JM McGree, K Mengersen, S Richardson, ... Statistical Science. 2017 Aug; 32(3): 385–404 |
2017 |
|
|
MA Ansari, V Pedergnana, CLC Ip, A Magri, A Von Delft, D Bonsall, ... Nature genetics 49 (5), 666 |
2017 |
|
|
Encrypted accelerated least squares regression PM Esperança, LJM Aslett, CC Holmes AISTATS 2017: The 20th International Conference on Artificial Intelligence and Statistics, 54, 334-343. |
2017 |
|
|
A note on statistical repeatability and study design for high‐throughput assays G Nicholson, C Holmes Statistics in medicine 36 (5), 790-798 |
2017 |
|
|
Bayesian Boolean Matrix Factorisation T Rukat, CC Holmes, MK Titsias, C Yau arXiv preprint arXiv:1702.06166 |
2017 |
This is to appear in ICML |
|
Digital Analysis of Tumour Microarchitecture as an Independent Prognostic Tool in Breast Cancer I Roxanis, R Colling, EA Rakha, A Green, J Rittscher, RC Conceicao, ... LABORATORY INVESTIGATION 97, 68A-68A |
2017 |
|
|
Assigning a value to a power likelihood in a general Bayesian model C Holmes, S Walker Biometrika, Volume 104, Issue 2, 497-503 |
2017 |
|
|
QF Wills, E Mellado-Gomez, R Nolan, D Warner, E Sharma, J Broxholme, ... BMC genomics 18 (1), 53 |
2017 |
|
|
On Markov chain Monte Carlo Methods for Tall Data A Doucet, CC Holmes, R Bardenet |
2017 |
|
|
Rejoinder: Approximate Models and Robust Decisions J Watson, C Holmes |
2017 |
|
|
Transcriptome Deconvolution of Heterogeneous Tumor Samples with Immune Infiltration Z Wang, JS Morris, S Cao, J Ahn, R Liu, S Tyekucheva, B Li, W Lu, X Tang, ... bioRxiv, 146795 |
2017 |
|
|
AR Taylor, JA Flegg, CC Holmes, PJ Guérin, CH Sibley, MD Conrad, ... Open Forum Infectious Diseases, ofw229 |
2016 |
|
|
Z Wang, JS Morris, CC Holmes, J Ahn, B Li, W Lu, X Tang, II Wistuba, ... Cancer Research 76 (14 Supplement), 2413-2413 |
2016 |
|
|
A general framework for updating belief distributions PG Bissiri, CC Holmes, SG Walker Journal of the Royal Statistical Society: Series B (Statistical Methodology) |
2016 |
|
|
Statistical inference in hidden Markov models using k-segment constraints MK Titsias, CC Holmes, C Yau Journal of the American Statistical Association 111 (513), 200-215 |
2016 |
|
|
S Filippi, CC Holmes, LE Nieto-Barajas Electronic Journal of Statistics 10 (2), 3338-3354 |
2016 |
|
|
Scalable Bayesian nonparametric regression via a Plackett-Luce model for conditional ranks T Gray-Davies, CC Holmes, F Caron Electronic Journal of Statistics 10 (2), 1807-1828 |
2016 |
|
|
A Bayesian nonparametric approach to testing for dependence between random variables S Filippi, CC Holmes Bayesian Analysis 12 (4), 919-938 |
2016 |
|
|
Approximate models and robust decisions J Watson, C Holmes Statistical Science 31 (4), 465-489 |
2016 |
|
|
AC Daly, DJ Gavaghan, C Holmes, J Cooper Royal Society open science 2 (12), 150499 |
2015 |
|
|
RGPM van Stiphout, L Winchester, SA Haider, J Ragoussis, AL Harris, ... Cancer Research 75 (22 Supplement 2), B1-56-B1-56 |
2015 |
|
|
MH de Angelis, G Nicholson, M Selloum, JK White, H Morgan, ... Nature genetics 47 (9), 969-978 |
2015 |
|
|
Encrypted statistical machine learning: new privacy preserving methods LJM Aslett, PM Esperança, CC Holmes arXiv preprint arXiv:1508.06845 |
2015 |
|
|
A review of homomorphic encryption and software tools for encrypted statistical machine learning LJM Aslett, PM Esperança, CC Holmes arXiv preprint arXiv:1508.06574 |
2015 |
|
|
Factors influencing success of clinical genome sequencing across a broad spectrum of disorders JC Taylor, HC Martin, S Lise, J Broxholme, JB Cazier, A Rimmer, ... Nature genetics 47 (7), 717-726 |
2015 |
|
|
Robust Linear Models for Cis-eQTL Analysis M Rantalainen, CM Lindgren, CC Holmes PloS one 10 (5), e0127882 |
2015 |
|
|
On Markov chain Monte Carlo methods for tall data R Bardenet, A Doucet, C Holmes arXiv preprint arXiv:1505.02827 |
2015 |
|
|
Two-sample Bayesian nonparametric hypothesis testing CC Holmes, F Caron, JE Griffin, DA Stephens Bayesian Analysis 10 (2), 297-320 |
2015 |
|
|
SJL Knight, R Clifford, P Robbe, SDC Ramos, A Burns, AT Timbs, ... Blood 124 (21), 3315-3315 |
2014 |
|
|
RM Clifford, P Robbe, S Weller, AT Timbs, M Titsias, A Burns, M Cabes, ... Blood 124 (21), 1942-1942 |
2014 |
|
|
Erythrocytosis associated with a novel missense mutation in the BPGM gene N Petousi, RR Copley, TRJ Lappin, SE Haggan, CM Bento, H Cario, ... haematologica 99 (10), e201-e204 |
2014 |
|
|
KE Pinnick, G Nicholson, KN Manolopoulos, SE McQuaid, P Valet, ... Diabetes, DB_140385 |
2014 |
|
|
AR Taylor, JA Flegg, SL Nsobya, A Yeka, MR Kamya, PJ Rosenthal, ... Malaria journal 13 (1), 102 |
2014 |
|
|
An adaptive subsampling approach for MCMC inference in large datasets R Bardenet, A Doucet, C Holmes Proceedings of The 31st International Conference on Machine Learning, 405-413 |
2014 |
|
|
Towards scaling up Markov chain Monte Carlo: an adaptive subsampling approach R Bardenet, A Doucet, C Holmes Proceedings of the 31st International Conference on Machine Learning (ICML ... |
2014 |
|
|
Nonlinear estimation and classification DD Denison, MH Hansen, CC Holmes, B Mallick, B Yu Springer Science & Business Media |
2013 |
|
|
D Mouradov, E Domingo, P Gibbs, RN Jorissen, S Li, PY Soo, L Lipton, ... The American journal of gastroenterology 108 (11), 1785-1793 |
2013 |
|
|
Integrative network-based Bayesian analysis of diverse genomics data W Wang, V Baladandayuthapani, CC Holmes, KA Do BMC bioinformatics 14 (13), S8 |
2013 |
|
|
Statistical estimation of malaria genotype frequencies: a Bayesian approach A Taylor, J Flegg, G Dorsey, P Guerin, C Holmes Tropical Medicine & International Health 18, 67 |
2013 |
|
|
Single-cell gene expression analysis reveals genetic associations masked in whole-tissue experiments QF Wills, KJ Livak, AJ Tipping, T Enver, AJ Goldson, DW Sexton, ... Nature biotechnology 31 (8), 748-752 |
2013 |
|
|
NucleoFinder: a statistical approach for the detection of nucleosome positions J Becker, C Yau, JM Hancock, CC Holmes Bioinformatics 29 (6), 711-716 |
2013 |
|
|
C Palles, JB Cazier, KM Howarth, E Domingo, AM Jones, P Broderick, ... Nature genetics 45 (2), 136-144 |
2013 |
|
|
Use of multivariate analysis to suggest a new molecular classification of colorectal cancer E Domingo, R Ramamoorthy, D Oukrif, D Rosmarin, M Presz, H Wang, ... The Journal of pathology 229 (3), 441-448 |
2013 |
|
|
Methods for qPCR gene expression profiling applied to 1440 lymphoblastoid single cells KJ Livak, QF Wills, AJ Tipping, K Datta, R Mittal, AJ Goldson, DW Sexton, ... Methods 59 (1), 71-79 |
2013 |
|
|
C Palles, J Cazier, K Howarth, E Domingo, A Jones, P Broderick, Z Kemp, ... Nature Genet 45, 136-144 |
2013 |
|
|
A decision-theoretic approach for segmental classification C Yau, CC Holmes The Annals of Applied Statistics 7 (3), 1814-1835 |
2013 |
|
|
A Novel Test for Gene-Ancestry Interactions in Genome-Wide Association Data JL Davies, JB Cazier, MG Dunlop, RS Houlston, IP Tomlinson, ... PloS one 7 (12), e48687 |
2012 |
|
|
JB Cazier, CC Holmes, J Broxholme Bioinformatics 28 (22), 2981-2982 |
2012 |
|
|
A Bayesian model for estimating with-in host P. falciparum haplotype frequencies AR Taylor, JA Flegg, PJ Guerin, C Roper, C Holmes Malaria Journal 11 (S1), P36 |
2012 |
|
|
Reprioritizing Genetic Associations in Hit Regions Using LASSO‐Based Resample Model Averaging W Valdar, J Sabourin, A Nobel, CC Holmes Genetic epidemiology 36 (5), 451-462 |
2012 |
|
|
F Caron, C Holmes, E Rio |
2012 |
|
|
F Caron, C Holmes, E Rio INRIA |
2012 |
|
|
nEASE: a method for gene ontology subclassification of high-throughput gene expression data TW Chittenden, EA Howe, JM Taylor, JC Mar, MJ Aryee, H Gómez, ... Bioinformatics 28 (5), 726-728 |
2012 |
|
|
JL Min, G Nicholson, I Halgrimsdottir, K Almstrup, A Petri, A Barrett, ... PLoS Genet 8 (2), e1002505 |
2012 |
|
|
SJL Knight, C Yau, R Clifford, AT Timbs, ES Akha, HM Dreau, A Burns, ... Leukemia 26 (7) |
2012 |
|
|
Bayesian sparsity-path-analysis of genetic association signal using generalized t priors A Lee, F Caron, A Doucet, C Holmes Statistical applications in genetics and molecular biology 11 (2), 1-29 |
2012 |
|
|
Variance decomposition of protein profiles from antibody arrays using a longitudinal twin model BS Kato, G Nicholson, M Neiman, M Rantalainen, CC Holmes, A Barrett, ... Proteome science 9 (1), 73 |
2011 |
|
|
M Rantalainen, BM Herrera, G Nicholson, R Bowden, QF Wills, JL Min, ... PloS one 6 (11), e27338 |
2011 |
|
|
J Ciampa, M Yeager, K Jacobs, MJ Thun, S Gapstur, D Albanes, J Virtamo, ... Human heredity 72 (3), 182-193 |
2011 |
|
|
Accounting for control mislabeling in case–control biomarker studies M Rantalainen, CC Holmes Journal of proteome research 10 (12), 5562-5567 |
2011 |
|
|
G Nicholson, M Rantalainen, JV Li, AD Maher, D Malmodin, KR Ahmadi, ... PLoS genetics 7 (9), e1002270 |
2011 |
|
|
Bayesian hierarchical mixture modeling to assign copy number from a targeted CNV array N Cardin, C Holmes, P Donnelly, J Marchini Genetic epidemiology 35 (6), 536-548 |
2011 |
|
|
Stochastic boosting algorithms A Jasra, CC Holmes Statistics and Computing 21 (3), 335-347 |
2011 |
|
|
C Yau, C Holmes Bayesian analysis (Online) 6 (2), 329 |
2011 |
|
|
JG Ciampa, C Holmes, N Chatterjee Cancer Research 71 (8 Supplement), 2747-2747 |
2011 |
|
|
A Drong, G Nicholson, M Schuster, F Karpe, MI McCarthy, CC Holmes, ... |
2011 |
|
|
C Holmes, L Held Bayesian Analysis 6 (2), 357-358 |
2011 |
|
|
Bayesian non‐parametric hidden Markov models with applications in genomics C Yau, O Papaspiliopoulos, GO Roberts, C Holmes Journal of the Royal Statistical Society: Series B (Statistical Methodology ... |
2011 |
|
|
Human metabolic profiles are stably controlled by genetic and environmental variation G Nicholson, M Rantalainen, AD Maher, JV Li, D Malmodin, KR Ahmadi, ... Molecular systems biology 7 (1), 525 |
2011 |
|
|
Therapeutic implications of GIPC1 silencing in cancer TW Chittenden, J Pak, R Rubio, H Cheng, K Holton, N Prendergast, ... PloS one 5 (12), e15581 |
2010 |
|
|
A Timbs, S Knight, E SadighiAkha, A Burns, H Dreau, AM Hewitt, C Hatton, ... Blood 116 (21), 3590-3590 |
2010 |
|
|
IM Heid, AU Jackson, JC Randall, TW Winkler, L Qi, V Steinthorsdottir, ... Nature genetics 42 (11), 949-960 |
2010 |
|
|
Elusive copy number variation in the mouse genome A Agam, B Yalcin, A Bhomra, M Cubin, C Webber, C Holmes, J Flint, ... PLoS One 5 (9), e12839 |
2010 |
|
|
C Yau, D Mouradov, RN Jorissen, S Colella, G Mirza, G Steers, A Harris, ... Genome biology 11 (9), R92 |
2010 |
|
|
A hierarchical Bayesian framework for constructing sparsity-inducing priors A Lee, F Caron, A Doucet, C Holmes arXiv preprint arXiv:1009.1914 |
2010 |
|
|
TG Clark, SG Campino, E Anastasi, S Auburn, YY Teo, K Small, ... Bioinformatics |
2010 |
|
|
JL Davies, J Hein, CC Holmes Genetic epidemiology 34 (4), 299-308 |
2010 |
|
|
Q Zhou, AKK Ching, WKC Leung, C Szeto, SM Ho, CC Holmes, Y Yuan, ... Cancer Research 70 (8 Supplement), 4423-4423 |
2010 |
|
|
Computational issues arising in Bayesian nonparametric hierarchical models J Griffin, C Holmes Bayesian Nonparametrics 28, 208 |
2010 |
|
|
An invitation to Bayesian nonparametrics NL Hjort, C Holmes, P Müller, SG Walker Bayesian Nonparametrics 28, 1 |
2010 |
|
|
NL Hjort, C Holmes, P Müller, SG Walker Cambridge University Press |
2010 |
|
|
N Craddock, ME Hurles, N Cardin, RD Pearson, V Plagnol, S Robson, ... Nature 464 (7289), 713-720 |
2010 |
|
|
M Rantalainen, BM Herrera, G Nicholson, QF Wills, R Bowden, MJ Neville, ... |
2010 |
|
|
Bayesian nonparametrics. Cambridge series in statistical and probabilistic mathematics NL Hjort, C Holmes, P Müller, SG Walker Cambridge: Cambridge Univ. Press. Mathematical Reviews (MathSciNet): MR2722987 |
2010 |
|
|
A Lee, C Yau, MB Giles, A Doucet, CC Holmes Journal of Computational and Graphical Statistics 19 (4), 769-789 |
2010 |
|
|
MA Suchard, C Holmes, M West Bulletin of the International Society for Bayesian Analysis 17 (1), 12-16 |
2010 |
|
|
A boosting approach to structure learning of graphs with and without prior knowledge S Anjum, A Doucet, CC Holmes Bioinformatics 25 (22), 2929-2936 |
2009 |
|
|
FE Mackenzie, A Parker, NJ Parkinson, PL Oliver, D Brooker, P Underhill, ... Genes, Brain and Behavior 8 (7), 699-713 |
2009 |
|
|
Mapping in structured populations by resample model averaging W Valdar, CC Holmes, R Mott, J Flint Genetics 182 (4), 1263-1277 |
2009 |
|
|
Reply to Wirth et al.: In vivo profiles show continuous variation between 2 cellular populations JE Lemieux, A Feller, CC Holmes, CI Newbold Proceedings of the National Academy of Sciences 106 (27), E71-E72 |
2009 |
|
|
A Antonyuk, C Holmes Genetic epidemiology 33 (5), 371-378 |
2009 |
|
|
JW Klingelhoefer, L Moutsianas, C Holmes Bioinformatics 25 (13), 1594-1601 |
2009 |
|
|
JE Lemieux, N Gomez-Escobar, A Feller, C Carret, A Amambua-Ngwa, ... Proceedings of the National Academy of Sciences 106 (18), 7559-7564 |
2009 |
|
|
Phylogenetic inference under recombination using Bayesian stochastic topology selection A Webb, JM Hancock, CC Holmes Bioinformatics 25 (2), 197-203 |
2009 |
|
|
Antithetic methods for gibbs samplers C Holmes, A Jasra Journal of Computational and Graphical Statistics 18 (2), 401-414 |
2009 |
|
|
Beyond toplines: Heterogeneous treatment effects in randomized experiments A Feller, CC Holmes Unpublished manuscript, Oxford University |
2009 |
|
|
E Giannoulatou, C Yau, S Colella, J Ragoussis, CC Holmes Bioinformatics 24 (19), 2209-2214 |
2008 |
|
|
Key issues in conducting a meta-analysis of gene expression microarray datasets A Ramasamy, A Mondry, CC Holmes, DG Altman PLoS Med 5 (9), e184 |
2008 |
|
|
TW Chittenden, EA Howe, AC Culhane, R Sultana, JM Taylor, C Holmes, ... Genomics 91 (6), 508-511 |
2008 |
|
|
Interacting sequential Monte Carlo samplers for trans-dimensional simulation A Jasra, A Doucet, DA Stephens, CC Holmes Computational Statistics & Data Analysis 52 (4), 1765-1791 |
2008 |
|
|
CNV discovery using SNP genotyping arrays C Yau, CC Holmes Cytogenetic and genome research 123 (1-4), 307-312 |
2008 |
|
|
Population-based reversible jump Markov chain Monte Carlo A Jasra, DA Stephens, CC Holmes Biometrika, 787-807 |
2007 |
|
|
CC Holmes, A Pintore Bayesian statistics 8: proceedings of the eighth Valencia International ... |
2007 |
|
|
On population-based simulation for static inference A Jasra, DA Stephens, CC Holmes Statistics and Computing 17 (3), 263-279 |
2007 |
|
|
Mcmc Methods for Bayesian Variable Selection in Large-scale Genomic Applications M Zucknick, C Holmes, S Richardson Annals of Human Genetics 71 (4), 558-559 |
2007 |
|
|
Flexible threshold models for modelling interest rate volatility P Dellaportas, DGT Denison, C Holmes Econometric reviews 26 (2-4), 419-437 |
2007 |
|
|
S Colella, C Yau, JM Taylor, G Mirza, H Butler, P Clouston, AS Bassett, ... Nucleic Acids Research 35 (6), 2013-2025 |
2007 |
|
|
Bayesian nonparametric calibration with applications in spatial epidemiology JE Griffin, CC Holmes Technical Report, Institute of Mathematics, Statistics and Actuarial Science ... |
2007 |
|
|
Integrating 3D information from thermochronological data over unknown spatial scales K Gallagher, J Stephenson, R Brown, C Holmes Geophysical Research Abstracts 9, 09015 |
2007 |
|
|
LJ Astle, CC Holmes, DJ Balding WILEY-LISS 31 (6), 605-605 |
2007 |
|
|
J Stephenson, K Gallagher, C Holmes Geochimica et Cosmochimica Acta 70 (20), 5183-5200 |
2006 |
|
|
Putting the data to work—strategies for modelling multiple samples in multiple dimensions K Gallagher, J Stephenson, C Holmes, R Brown Geochimica et Cosmochimica Acta 70 (18), A190 |
2006 |
|
|
A new approach to mixture modelling for geochronology K Gallagher, A Jasra, D Stephens, C Holmes Geochimica et Cosmochimica Acta 70 (18), A190 |
2006 |
|
|
Bayesian mixture modelling in geochronology via Markov chain Monte Carlo A Jasra, DA Stephens, K Gallagher, CC Holmes Mathematical geology 38 (3), 269-300 |
2006 |
|
|
A quantitative study of gene regulation involved in the immune response of anopheline mosquitoes NA Heard, CC Holmes, DA Stephens Journal of the American Statistical Association 101 (473), 18-29 |
2006 |
|
|
Spatially adaptive smoothing splines A Pintore, P Speckman, CC Holmes Biometrika, 113-125 |
2006 |
|
|
J Stephenson, K Gallagher, CC Holmes Earth and Planetary Science Letters 241 (3), 557-570 |
2006 |
|
|
V Baladandayuthapani, CC Holmes, BK Mallick, RJ Carroll |
2006 |
|
|
Analysis of geochronological data with measurement error using Bayesian mixtures A Jasra, DA Stephens, K Gallagher, CC Holmes Mathematical Geology 38, 269-300 |
2006 |
|
|
Modeling nonlinear gene interactions using Bayesian MARS V Baladandayuthapani, CC Holmes, BK Mallick, RJ Carroll Bayesian Inference for Gene Expression and Proteomics. Cambridge University ... |
2006 |
|
|
Modulation of the BK channel by estrogens: examination at single channel level H De Wet, M Allen, C Holmes, M Stobbart, JD Lippiat, H De Wet, M Allen, ... Molecular membrane biology 23 (5), 420-429 |
2006 |
|
|
Bayesian auxiliary variable models for binary and multinomial regression CC Holmes, L Held Bayesian analysis 1 (1), 145-168 |
2006 |
|
|
Bayesian prediction via partitioning CC Holmes, DGT Denison, S Ray, BK Mallick Journal of Computational and Graphical Statistics 14 (4), 811-830 |
2005 |
|
|
NA Heard, CC Holmes, DA Stephens, DJ Hand, G Dimopoulos Proceedings of the National Academy of Sciences of the United States of ... |
2005 |
|
|
Low temperature thermochronology and modeling strategies for multiple samples 1: Vertical profiles K Gallagher, J Stephenson, R Brown, C Holmes, P Fitzgerald Earth and Planetary Science Letters 237 (1), 193-208 |
2005 |
|
|
Analyzing nonstationary spatial data using piecewise Gaussian processes HM Kim, BK Mallick, CC Holmes Journal of the American Statistical Association 100 (470), 653-668 |
2005 |
|
|
Markov chain Monte Carlo methods and the label switching problem in Bayesian mixture modeling A Jasra, CC Holmes, DA Stephens Statistical Science, 50-67 |
2005 |
|
|
All systems GO for understanding mouse gene function C Holmes, SDM Brown The Scientist 19 (1), 20.1-20.1 |
2005 |
|
|
A statistical technique for modelling non-stationary spatial processes J Stephenson, C Holmes, K Gallagher, A Pintore Geostatistics Banff 2004, 125-134 |
2005 |
|
|
A dimension-reduction approach for spectral tempering using empirical orthogonal functions A Pintore, CC Holmes Geostatistics Banff 2004, 1007-1015 |
2005 |
|
|
Exploiting 3D spatial sampling in inverse modeling of thermochronological data K Gallagher, J Stephenson, R Brown, C Holmes, P Ballester Reviews in mineralogy and geochemistry 58 (1), 375-387 |
2005 |
|
|
All systems GO for understanding mouse gene function C Holmes, SDM Brown Journal of biology 3 (5), 20 |
2004 |
|
|
J Stephenson, K Gallagher, CC Holmes Geological Society, London, Special Publications 239 (1), 195-209 |
2004 |
|
|
Spatially adaptive non-stationary covariance functions via spatially adaptive spectra A Pintore, C Holmes http:\\ www. stats. ox. ac. uk cholmes\ Reports\ spectral tempering. pdf |
2004 |
|
|
Generalized nonlinear modeling with multivariate free-knot regression splines CC Holmes, BK Mallick Journal of the American Statistical Association 98 (462), 352-368 |
2003 |
|
|
Likelihood inference in nearest-neighbour classification models CC Holmes, NM Adams Biometrika, 99-112 |
2003 |
|
|
Generalized monotonic regression using random change points CC Holmes, NA Heard Statistics in Medicine 22 (4), 623-638 |
2003 |
|
|
CC Holmes, DGT Denison Machine Learning 50 (3), 279-301 |
2003 |
|
|
Gauss mixture quantization: clustering Gauss mixtures RM Gray, DD Denison, MH Hansen, CC Holmes, B Mallick, B Yu Nonlinear Estimation and Classification 1003, 189-212 |
2003 |
|
|
Bayesian free knot polynomial splines of random order R Graziani, C Holmes Università commerciale Luigi Bocconi |
2003 |
|
|
Perfect simulation for Bayesian curve and surface fitting CC Holmes, BK Mallick Preprint from www. stat. tamu. edu/~ bmallick/papers/perf. ps |
2003 |
|
|
Classification with bayesian MARS CC Holmes, DGT Denison Machine Learning 50 (1), 159-173 |
2003 |
|
|
Efficient simulation of Bayesian logistic regression models C Holmes, L Knorr-Held Discussion papers/Sonderforschungsbereich 386 der Ludwig-Maximilians ... |
2003 |
|
|
Accounting for model uncertainty in seemingly unrelated regressions CC Holmes, DGT Denison, BK Mallick Journal of Computational and Graphical Statistics 11 (3), 533-551 |
2002 |
|
|
Perfect simulation involving functionals of a Dirichlet process A Guglielmi, CC Holmes, SG Walker Journal of Computational and Graphical Statistics 11 (2), 306-310 |
2002 |
|
|
J Ferreira, DGT Denison, CC Holmes |
2002 |
|
|
A probabilistic nearest neighbour method for statistical pattern recognition CC Holmes, NM Adams Journal of the Royal Statistical Society: Series B (Statistical Methodology ... |
2002 |
|
|
DGT Denison, NM Adams, CC Holmes, DJ Hand Computational statistics & data analysis 38 (4), 475-485 |
2002 |
|
|
Perfect sampling for the wavelet reconstruction of signals C Holmes, DGT Denison IEEE Transactions on Signal Processing 50 (2), 337-344 |
2002 |
|
|
Minimum-entropy data partitioning using reversible jump Markov chain Monte Carlo SJ Roberts, C Holmes, D Denison IEEE Transactions on Pattern Analysis and Machine Intelligence 23 (8), 909-914 |
2001 |
|
|
Bayesian partitioning for estimating disease risk DGT Denison, CC Holmes Biometrics 57 (1), 143-149 |
2001 |
|
|
Minimum-entropy data clustering using reversible jump markov chain monte carlo S Roberts, C Holmes, D Denison Artificial Neural Networks—ICANN 2001, 103-110 |
2001 |
|
|
Bayesian regression with multivariate linear splines CC Holmes, BK Mallick Journal of the Royal Statistical Society: Series B (Statistical Methodology ... |
2001 |
|
|
Bayesian wavelet networks for nonparametric regression CC Holmes, BK Mallick IEEE transactions on neural networks 11 (1), 27-35 |
2000 |
|
|
Generalised nonlinear modelling with multivariate smoothing splines CC Holmes, BK Mallick Unpublished manuscript, Statistics Section, Department of Mathematics ... |
1999 |
|
|
Bayesian wavelet analysis with a model complexity prior CC Holmes, DGT Denison Bayesian statistics 6, 769-776 |
1999 |
|
|
Bayesian radial basis functions of variable dimension CC Holmes, BK Mallick Neural Computation 10 (5), 1217-1233 |
1998 |
|
|
Model-based geostatistics-Discussion R Webster, A Lawson, C Glasbey, G Horgan, D Elston, G Host, ... |
1998 |
|