BNPgraph package: demo_graph
This Matlab script shows how to sample an undirected graph from a generalized gamma process graph model and how to perform posterior inference.
For downloading the package and information on installation, visit the BNPgraph webpage.
Reference: F. Caron and E.B. Fox. Sparse graphs using exchangeable random measures. arXiv:1401.1137, 2014.
Author: François Caron, University of Oxford
Tested on Matlab R2014a.
Contents
General settings
% Add paths addpath('./GGP'); addpath('./utils'); % Set the fontsize set(0,'DefaultAxesFontSize',14) % Set the seed rng('default')
Simulation of a GGP graph
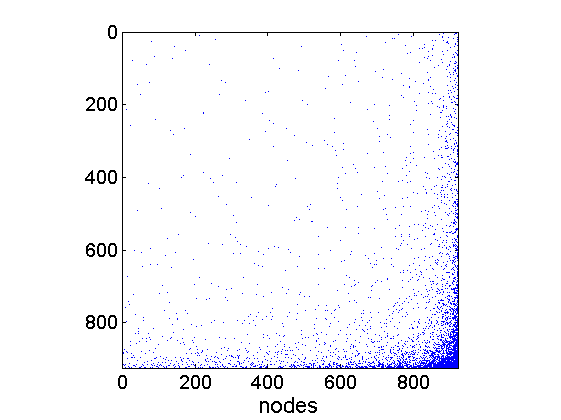
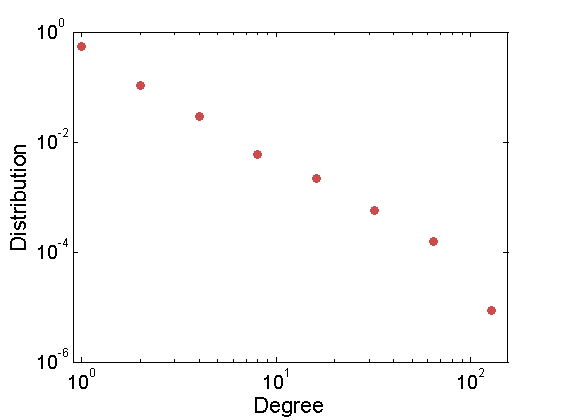
% Sample graph alpha = 50; tau = 1; sigma = .5; obj = graphmodel('GGP', alpha, sigma, tau) [G, wtrue, wtrue_rem] = graphrnd(obj,1e-6); % Plot the adjacency matrix figure; spy(G) xlabel('nodes', 'fontsize', 16) % Plot the empirical degree distribution deg = sum(G); n = size(G, 1); figure; h = plot_degree(G, 'o'); set(h, 'markersize', 6, 'color', [.8, .3, .3], 'markerfacecolor', [.8, .3, .3]); xlim([.9, max(deg)]);
obj =
graphmodel with properties:
name: 'Generalized gamma process'
type: 'GGP'
param: [1x1 struct]
typegraph: 'undirected'
MCMC inference for the GGP graph
% Prior graph model hyper_alpha =[0,0]; % Improper prior on alpha hyper_tau = [0,0]; % improper prior on tau hyper_sigma = [0, 0]; % Improper prior on sigma objprior = graphmodel('GGP', hyper_alpha, hyper_sigma, hyper_tau); % Run MCMC niter = 20000; nburn = 0; nadapt = niter/4; thin = 1; nchains = 3; verbose = false; objmcmc = graphmcmc(objprior, niter, nburn, thin, nadapt, nchains); objmcmc = graphmcmcsamples(objmcmc, G, verbose);
-----------------------------------
MCMC chain 1/3
-----------------------------------
Start MCMC for GGP graphs
Nb of nodes: 923 - Nb of edges: 2211
Number of iterations: 20000
Estimated computation time: 0 hour(s) 1 minute(s)
Estimated end of computation: 01-May-2015 11:33:05
-----------------------------------
-----------------------------------
End MCMC for GGP graphs
Computation time: 0 hour(s) 1 minute(s)
End of computation: 01-May-2015 11:32:28
-----------------------------------
-----------------------------------
MCMC chain 2/3
-----------------------------------
Start MCMC for GGP graphs
Nb of nodes: 923 - Nb of edges: 2211
Number of iterations: 20000
Estimated computation time: 0 hour(s) 1 minute(s)
Estimated end of computation: 01-May-2015 11:33:19
-----------------------------------
-----------------------------------
End MCMC for GGP graphs
Computation time: 0 hour(s) 1 minute(s)
End of computation: 01-May-2015 11:33:08
-----------------------------------
-----------------------------------
MCMC chain 3/3
-----------------------------------
Start MCMC for GGP graphs
Nb of nodes: 923 - Nb of edges: 2211
Number of iterations: 20000
Estimated computation time: 0 hour(s) 1 minute(s)
Estimated end of computation: 01-May-2015 11:34:17
-----------------------------------
-----------------------------------
End MCMC for GGP graphs
Computation time: 0 hour(s) 1 minute(s)
End of computation: 01-May-2015 11:33:48
-----------------------------------
Plot some summary statistics of the posterior
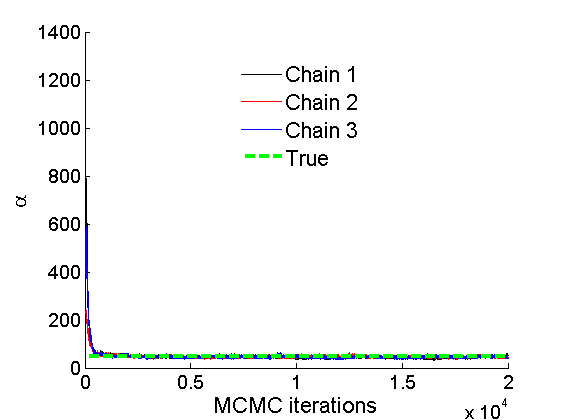
% Concatenate samples nsamples = length(objmcmc.samples(1).w_rem); [samples_all] = graphest(objmcmc, nsamples/2); [~, K] = size(samples_all.w); % Trace plots and posterior histograms of alpha, sigma, tau and w* col = {'k', 'r', 'b'}; variables = {'alpha', 'sigma', 'tau', 'w_rem'}; namesvar = {'\alpha', '\sigma', '\tau', 'w_*'}; truevalues = {obj.param.alpha, obj.param.sigma, obj.param.tau, wtrue_rem}; for i=1:length(variables) figure for k=1:nchains plot(thin:thin:niter, objmcmc.samples(k).(variables{i}), col{k}); hold on end plot(thin:thin:niter, truevalues{i}*ones(nsamples, 1), '--g', 'linewidth', 3); legend({'Chain 1','Chain 2', 'Chain 3', 'True'}, 'fontsize', 16, 'location', 'Best') legend boxoff xlabel('MCMC iterations', 'fontsize', 16); ylabel(namesvar{i}, 'fontsize', 16); box off xlim([0, niter]) figure hist(samples_all.(variables{i}), 30) hold on plot(truevalues{i}, 0, 'dg', 'markerfacecolor', 'g') xlabel(namesvar{i}, 'fontsize', 16); ylabel('Number of MCMC samples', 'fontsize', 16); end
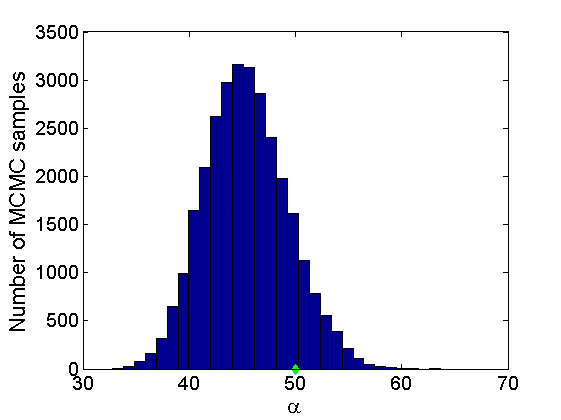
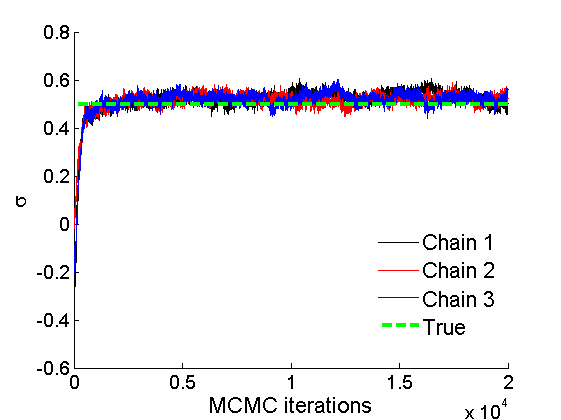
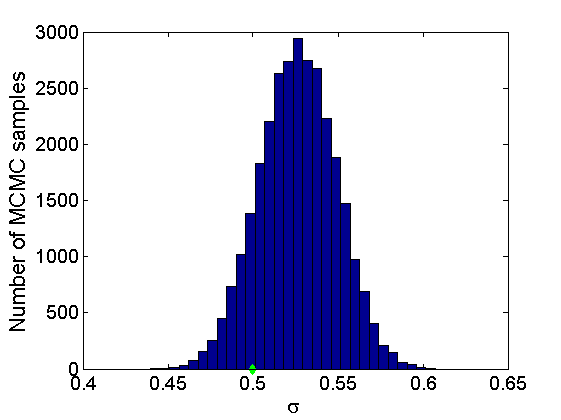
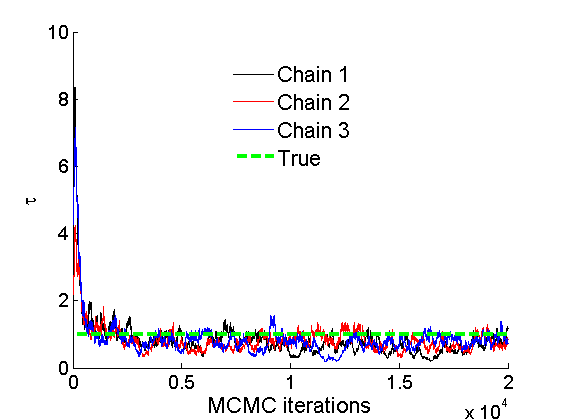
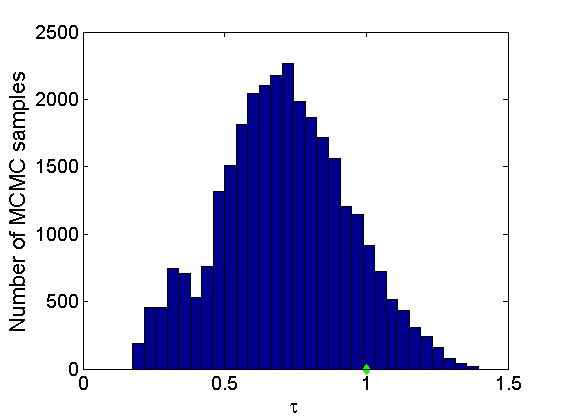
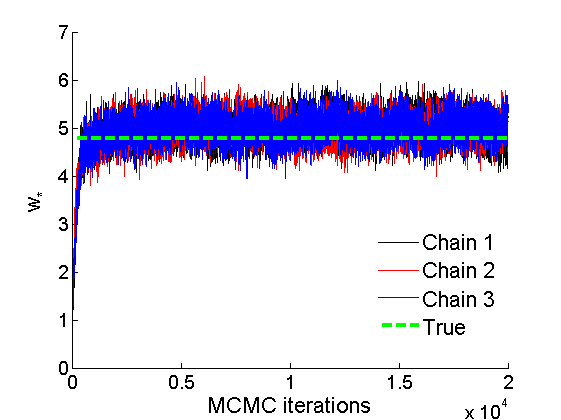
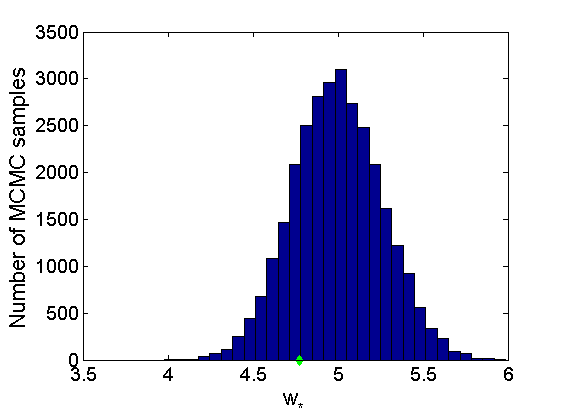
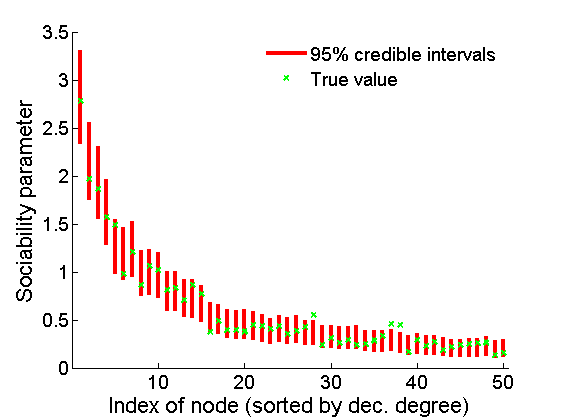
% Credible intervals for the weights [~, ind] = sort(sum(G), 'descend'); figure for k=1:min(K, 50) plot([k, k],... quantile(samples_all.w(:,ind(k)),[.025,.975]), 'r', ... 'linewidth', 3); hold on plot(k, wtrue(ind(k)), 'xg', 'linewidth', 2) end xlim([0.1,min(K, 50)+.5]) legend('95% credible intervals', 'True value') legend boxoff box off xlabel('Index of node (sorted by dec. degree)', 'fontsize', 16) ylabel('Sociability parameter', 'fontsize', 16)